Knowledge Base Contribution Guide
last updated at 2023-07-19Your contribution to the DataPLANT Knowledge Base is highly appreciated. This guide is intended to show you how to contribute new articles and tutorials or review and adapt parts of existing ones. For changes and suggestions, feel free to open a GitHub issue or pull request.
UserData Steward ModeTutorialBefore contributing to the knowledge base, you should have
☑️ a GitHub account and some routine with GitHub
☑️ an up-to-date version of Node.JS installed
☑️ an up-to-date version of .NET installed
💡 We recommend working with VS Code, with extensions for easy markdown editing and spell check.
💡 For a general introduction to writing markdown, see the markdown tutorial and references therein.
🚀 Feel free to contact us for Data Steward support.
The DataPLANT Knowledge Base is built on nfdi-web-components which fit markdown content into this "framework".
Please follow these steps to fork the knowledge base repository and clone your fork to your computer.
Fork the Knowledge Base Git repository.
- This creates a copy of the Knowledge Base repository in your own GitHub account.
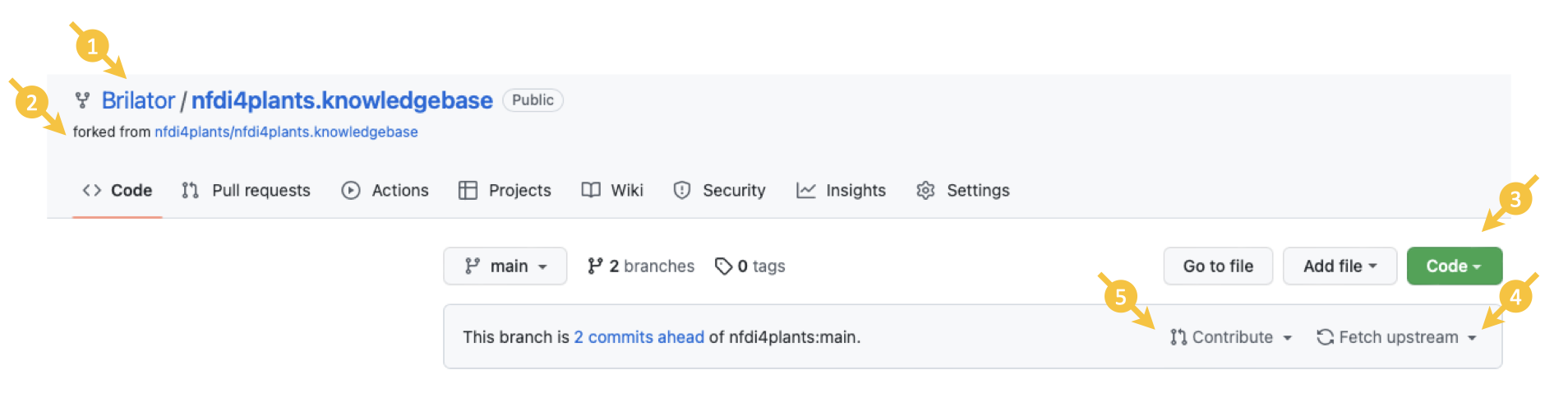
- In the top left, you will see that the repository is associated with your account (1) and forked from the main repository (2).
- You can either directly add or edit content using GitHub or clone (3) your repository to work on it locally.
- Be aware that your fork is not automatically updated, if the main repository updates. Make sure to update your fork regularly (especially before creating new content) by clicking "Fetch upstream" (4) in the top-right corner of your repository.
- If you cloned your repository locally, you also have to update the local clone (via "git pull").
You can work and make any changes in your own repository and commit + push them to your fork.
Once you want to submit those changes to the main repository, you can open a "pull request" by clicking "Contribute" (5) in the top-right corner.
Remember to "Fetch upstream" (4), if your fork is not up-to-date with the main repository.
If edited or added existing content, please assign the original author during your pull-request to review your changes.
- The github user name of the original author is stored in the yaml block on top under
author // github - During the pull-request mention the author via
@<author_github>.
⚠️ This is not a full-fledged GitHub tutorial. Please refer to available tutorials online or contact the Knowledge Base curators.
The following instructions allow you to test and see how your changes come into play and check whether everything renders correctly.
⚠️ It's highly recommended to frequently check your changes locally. Please do not produce and submit a lot of content without prior local testing.
- Download the repo.
- Run
dotnet tool restorein root directory. - Run
dotnet paket installin root directory. - Run
npm installin root directory.
💡 This needs to be done only once after cloning the repo.
- Run
npm run fornax. - Open the page in your browser http://127.0.0.1:8080/.
This will update the npm package nfdi4plants/web-components to latest, as well as update the nuget dependency Nfdi4Plants.Fornax to latest. Then it will bundle all npm (javascript) dependencies to a single bundle.js file with rollup.
This is part of the execution chain of npm run updatecomponents
Index created html files. Creates src/_public/pagefind folder. MUST be used after running npm run fornax at least once. Otherwise there will be no .html files to index.
Same as npm run index but starts local server to test search bar. Currently the only way to test search bar locally, but will not allow to track changes in markdown files like npm run fornax does (If you want to combine both a PR would be more than welcome).
Read more about authoring content in the fornax section of the nfdi4plants web components docs
⚠️ Please make sure to especially follow the markdown syntax section.
💡 The following sections refer to contents with layout: docs as used for articles and guides.
🚧 Contribution of slides (i.e. in folder src/docs/teaching-materials/ and sidebar section "Teaching Materials") is currently under construction. See the additional guide on slide decks.
- The source to all content shown at the public website https://nfdi4plants.org/nfdi4plants.knowledgebase/ resides in the folder
src/docs - All other files are mostly "backend", help to render the content to html and can safely be ignored by most contributors
- The
src/docsstructure (as of July 17th, 2023) looks like this
💡 For easier findability, we try to align the folder structure with how the content is presented in the sidebar of the public site. However, this is not always feasible or sensible (for reasons).
- Missing metadata block
- Missing or false required (
MUST) attribute in metadata block - Using a layout in metadata block that does not exist or is deprecated
- Wrong links
- to sidebar elements
- to images
Every markdown document with the YAML key layout: docs stored in /nfdi4plants.knowledgebase/src/docs or any subfolder
(except _ignored) will be rendered to html and become publicly available once pushed to the main repository.
- Anything inside the folder
_ignoredwill not be used to generate html pages. - This folder is intended for ideas and drafts that
- should not yet become available in the Knowledge Base
- should be under version control for discussion and reviewing
Although most markdown content will be rendered and published, it will not prominently be visible to all visitors.
It will only become visible in the sidebar once the article is linked in the respective sidebar
(typically add sidebar: _sidebars/mainSidebar.md)
- Literature / information references: additional bibliography block below
- External links (tools, sites, platforms): as hyper-link
- E.g. like this:
[CLICKABLE TEXT](https://daringfireball.net/projects/markdown/syntax#link) resulting in CLICKABLE TEXT - or like this e.g. in the FAQ.md file:
<a href="https://en.wikipedia.org/wiki/Hyperlink">CLICKABLE TEXT</a> resulting in: CLICKABLE TEXT
- E.g. like this:
- Knowledge Base cross-references: relative path to *.md document, BUT replace the
.mdfile extension with.html, as the markdown files are parsed to html.
We SHOULD always try to use relative paths, as they are easier to maintain.
Although useful, they need a bit more fine tuning for different purposes.
Relative paths in production are differently accessed than in development. One of the major issues with relative paths is that during development the pages are accessed by /, for example /docs/README.html.
Published they will be accessed by /nfdi4plants.knowledgebase (/nfdi4plants.knowledgebase/docs/README.html).
In the following, some ideas are described on how to deal with this:
Basic relative paths: A basic relative path looks like this:
[Test](/README.html).
By starting with/we implicitly say "start at host".
In development the same path will start withhttp://127.0.0.1:8080/, in production it will start withhttps://nfdi4plants.github.io/.
In production we also usehttps://nfdi4plants.org/nfdi4plants.knowledgebase/.Relative paths in sidebar: The sidebars actually checks if you are currently in
npm run fornaxmode. Therefore, you can use basic relative paths for this. (mainSidebar.md)For example
```Fundamentals # [Fundamentals](/docs/fundamentals/index.html) ```! /docs/fundamentals/index.htmlwill be parsed to/nfdi4plants.knowledgebase/docs/fundamentals/index.html.Relative paths from current file:
These might be easier to handle but need more maintenance as they MUST be changed when the folder/file structure changes, but they circumvent the basepath issue of "Basic relative paths". Examples:[Test](./ResearchDataManagement.html). Starting with./translates to "coming from the position of this file".- We can go up in the file hierarchy, like
[Test](./../docs/README.html). - For the link "./../docs/Home.html", this translates to "coming from the position of this file, go one folder higher and into the docs directory to find the Introduction which is named "Home" there."
Try to avoid deep structures by using no more than two headline levels, i.e.
```
## Headline level 2
### Headline level 3
```Headline level 1 is automatically generated from the article's title: .
Images can be linked
- via simple markdown logic:
 - via html:
<img src="path_to_image.png">
💡 Please store images in src/docs/img.
- The folder
src/docs/imgholds image files (preferably *.svg, *.png) or the original source file (.pptx, .drawio.svg) used to create the image file(s). - Avoid adding captions or links to figure files.
- If powerpoint was used to create a figure:
- The pptx stored in
src/docs/imgis supposed to be a file of only a single slide or slide sequence (i.e. consecutive slides, where one image builds on the previous) - The name of the resulting figure file (e.g. "FAIRprinciples.png") MUST be aligned with the source file (e.g. "FAIRprinciples.pptx")
- a consecutive number is suffixed to images from slide sequences (e.g. "FAIRprinciples_seq1.png", "FAIRprinciples_seq2.png")
- The pptx stored in
- Make sure you are allowed to add the content. If you reuse any content from other sources, make sure to reference the source and / or license, if applicable.
- In particular, when adding images that you did not create yourself, please clearly state this by
- adding the source to the
src/docs/img/_ImageIndex.md, - adding a link / license (however applicable) to the content (article, tutorial, slide deck, etc.) where you use the image, and
- reminding the knowledge base curators during your pull request.
- adding the source to the
File names:
- MUST use the article's title for the filename, e.g.: DataManagementPlan.md →
title: Data Management Plan - MUST NOT contain special characters
- MUST NOT contain spaces
- MUST use snake_case (lower_case_with_underscores) OR PascalCase (UpperCase)
⚠️ Changing file names (and paths) means changing URLs and can easily lead to dead links.
Once in a while, we (need to) restructure the /nfdi4plants.knowledgebase/src/docs folder a bit, which includes changes to file and folder names, i.e. producing dead links.
In this case, the least we can do, is to keep the knowledge base itself intact:
- Carefully check all markdown documents for cross-links to the original file name (this can easily be done via e.g. VS Code).
- Remember to check for both file endings
.mdand.html. - Replace with the new file name / path.
This however does not help for articles, tutorials, etc. that have been shared with the community (via mail, in slide decks, etc.). In this case, please leave a markdown with the original file name and path and just fill it with the following
This is not the most beautiful solution as it clutters the knowledge base, but it helps against user frustration and should be done for frequently shared articles. Adding a date helps us clean up those files after a certain time interval.
💡 If you're sharing content with the users, it might be best to only share the link to the knowledge base https://nfdi4plants.org/nfdi4plants.knowledgebase/, rather than a concrete link to a tutorial (e.g. https://nfdi4plants.org/nfdi4plants.knowledgebase/docs/guides/arcCommander_QuickStart.html).
We generally try to address users and readers directly. Use "you can", not "the user can" or "one can..."
British English
Note: If you work with Visual Studio Code - Check out the extension "Code Spell Checker" https://marketplace.visualstudio.com/items?itemName=streetsidesoftware.code-spell-checker with British English ("cSpell.language": "en-GB") support.