--- ## File Export - Click **save** button to download the file - file can also be exported as [ISA-JSON](https://isa-specs.readthedocs.io/en/latest/isajson.html#) file --- ## Re-use a protocol (process.json) 1. Import the empty `assays/metabolomics/isa.assay.xlsx)` workbook in Swate. 2. Navigate to *Templates* in the Navbar and scroll down to "Add template(s) from file." 3. Click <kbd>Upload protocol</kbd> 4. Select the file "swate_agilent_gc.json" from the demo data. 5. Click <kbd>Insert json</kbd> 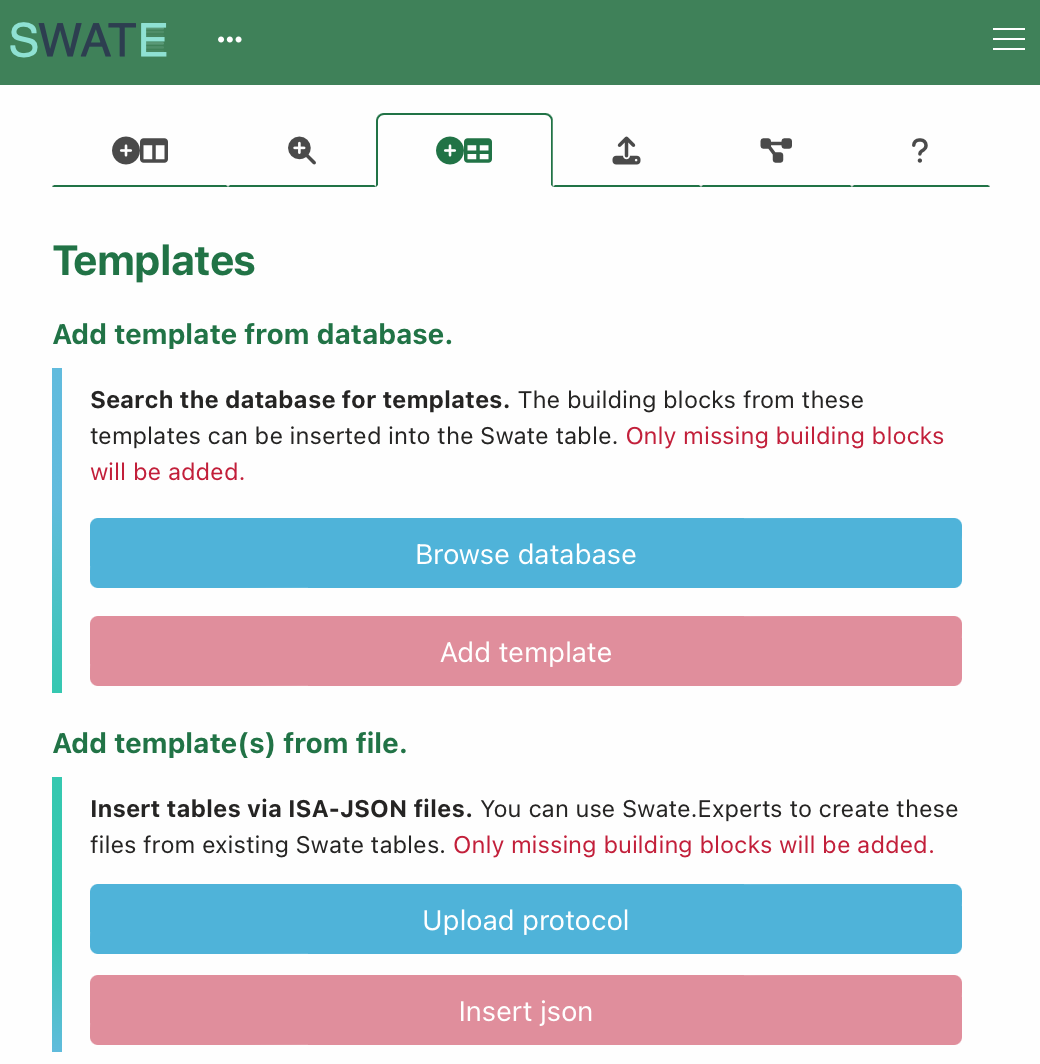 :bulb: This adds not only an empty template, but a filled out table with keys (headers) and values (cells). --- ## A small detour on "Excel Tables" Swate uses Excel's "table" feature to annotate workflows. Each table represents one *process* from input (e.g. plant leaf material) to output (e.g. leaf extract). Example workflows with three *processes* each: - Plant growth → sampling → extraction - Measured data files → statistical analysis → result files :bulb: Excel tables allow to group data that belongs together inside one sheet. This is not to be confused with a (work)sheet or workbook. > ```bash > workbook (e.g. "isa.assay.xlsx") > └─── worksheet (e.g. "plant_growth") > └─── table (e.g. "annotationTable") ---